Nalm6-Fluc-Neo/eGFP-Puro
| Species | Human |
| Cell Type | Lymphoma |
| Transgene | Firefly luciferase (Fluc) Enhanced green fluorescent protein (eGFP) |
| Selection Gene | Neomycin (neo) Puromycin resistance (puro) |
-
Description
Parental Nalm6 cells were transduced with 1) LV-Fluc-P2A-Neo (Imanis #LV011) encoding the firefly luciferase (Fluc) cDNA under the spleen focus-forming virus (SFFV) promoter and linked to the neomycin resistance gene (Neo) via a P2A cleavage peptide and 2) LV-eGFPPGK-Puro (Imanis #LV031) encoding the enhanced green fluorescent protein (eGFP) cDNA under the SFFV promoter and the puromycin resistance gene (Puro) under the phosphoglycerate kinase (PGK) promoter. A high Fluc and eGFP expressing population was generated by selection using G418 and puromycin followed by selection using a methylcellulose based semi-solid medium.
The parental Nalm6 cell line was authenticated and certified free of interspecies cross contamination by STR profiling.
This cell line has been tested for mycoplasma contamination and is certified mycoplasma free.
-
Reporter Gene Validation
In Vivo Imaging
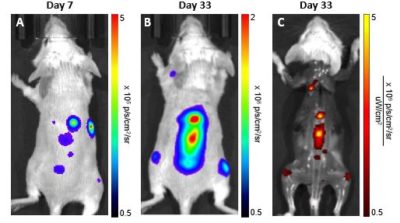 SCID beige mice were injected intraveneously with 1 x 106 Nalm6-Fluc-Neo/eGFP-Puro (CL150) cells. Bioluminescent (A&B) and fluorescent (C) imaging was performed at the indicated times.
SCID beige mice were injected intraveneously with 1 x 106 Nalm6-Fluc-Neo/eGFP-Puro (CL150) cells. Bioluminescent (A&B) and fluorescent (C) imaging was performed at the indicated times.Morphology
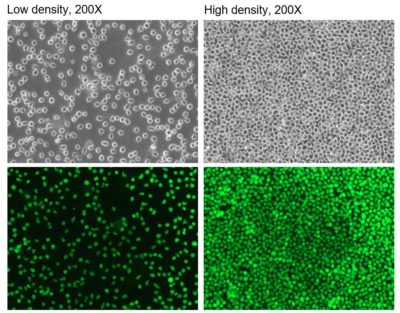 Low and high density photos taken at different times after thawing.
Low and high density photos taken at different times after thawing.Luciferase Expression
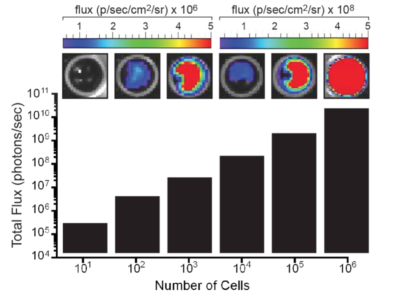
The indicated number of cells were placed in wells of a 96-well plate. After the addition of 3 mg/mL d-luciferin, the plate was immediately imaged using an IVIS Spectrum. The total flux (photons/sec) was plotted as a function of cell number.
eGFP Expression
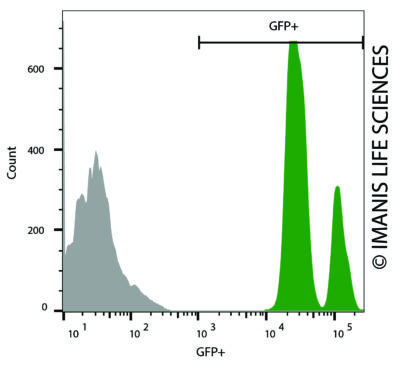
Nalm6-Fluc-Neo/eGFP-Puro (green) or isotype control (Nalm6-Fluc-Puro grey) cells were fixed with paraformaldehyde and analyzed by flow cytometry. Nalm6-FlucNeo/eGFP-Puro (green) or isotype control (Nalm6-Fluc-Puro grey) cells were fixed with paraformaldehyde and analyzed by flow cytometry.
-
Growth Conditions
Complete Growth Medium: RPMI-1640 Medium (RPMI) containing 10 mM HEPES supplemented with 10% FBS, 1% Penicillin/Streptomycin, 1 mg/mL G418, and 1 µg/mL puromycin.
The addition of puromycin to the complete growth medium maintains high reporter expression over continued passage of the cells. It is highly recommended, especially if the cells undergo multiple passages prior to being used for studies.
These cells should be grown in the indicated medium and subcultured as needed to maintain a density between 3 x 105 and 3 x 106 cells/mL. The cells can be passaged by dilution in fresh medium. Regular passaging using centrifugation is recommended to limit the amount of debris in cultures.
-
Usage Information
These cells are suitable for in vitro and in vivo experimentation.
Nalm6 cells form tumors and metastases at multiple sites, including the liver, lymph nodes, spleen, long bones, periodontal region, and bone marrow, upon implantation into immunocompromised mice1,2.
The Fluc transgene facilitates in vivo noninvasive bioluminescent imaging of implanted cells. eGFP is not recommended for whole animal in-live imaging. Rather, samples can be collected post mortem for analysis by conventional fluorescence microscopy or flow cytometry.
These cells were generated via lentiviral vector transduction. The lentiviral vector used for transduction was a self-inactivating (SIN) vector in which the viral enhancer and promoter have been deleted. Transcription inactivation of the LTR in the SIN provirus increases biosafety by preventing mobilization by replication competent viruses and enables regulated expression of the genes from the internal promoters without cis-acting effects of the LTR2. Nevertheless, all work with these cells should be performed under biosafety-level 2 (BSL2) conditions by trained personnel. Institutional requirements may permit handling of these cells under BSL1 conditions if certain criteria are met.
1Santos et al. Nature Med 2009 15: 338-344.
2Brentjens et al. Clin Cancer Res 2007 13: 5426-5435. -
Datasheet/COA
Lot Number IMP038